BWA-MEME
BWA-MEM emulated with a machine learning approach
Summary
The growing use of next-generation sequencing and enlarged sequencing throughput require efficient
short-read alignment, where seeding is one of the major performance bottlenecks. The key challenge in the seeding
phase is searching for exact matches of substrings of short reads in the reference DNA sequence. Existing algorithms, however, present limitations in performance due to their frequent memory accesses.
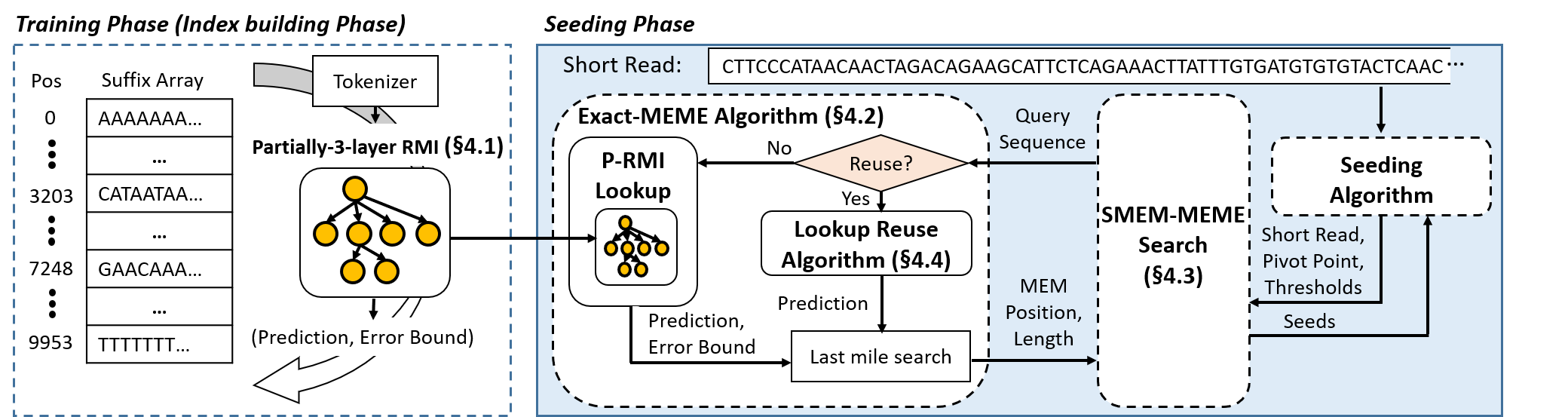
BWA-MEME is the first full-fledged short read alignment software that leverages learned
indices for solving the exact match search problem for efficient seeding. BWA-MEME is a practical and efficient
seeding algorithm based on a suffix array search algorithm that solves the challenges in utilizing learned indices for
SMEM search which is extensively used in the seeding phase. Our evaluation shows that BWA-MEME achieves up
to 3.45 speedup in seeding throughput over BWA-MEM2 by reducing the number of instructions by 4.60, memory accesses by 8.77 and LLC misses by 2.21, while ensuring the identical SAM output to BWA-MEM2.

